Next Generation Sequencing (NGS) is becoming a hot topic in diagnostics. But how is the data generated and processed so it can be used to develop diagnostic tests? We speak to Michael Schmid, founder of Genexa, a bioinformatics start-up which has worked with EU-based labs to solve these problems.
1) Bioinformatics plays a huge role in interpreting the data produced in NGS. Could you please describe the workflow behind NGS?
Before the advent of NGS, even “deciphering” the small and comparably simple genome sequences of viruses or bacteria was a very laborious task and could easily take months to years.
Sequencing the human genome was even more challenging. An international project to sequence the human genome in the 90s to early 2000s was performed by hundreds of scientists at about 20 universities and cost billions of dollars. Today, with the help of NGS we can achieve the same task of reading the human genome for a couple of thousand dollars and within days to weeks.
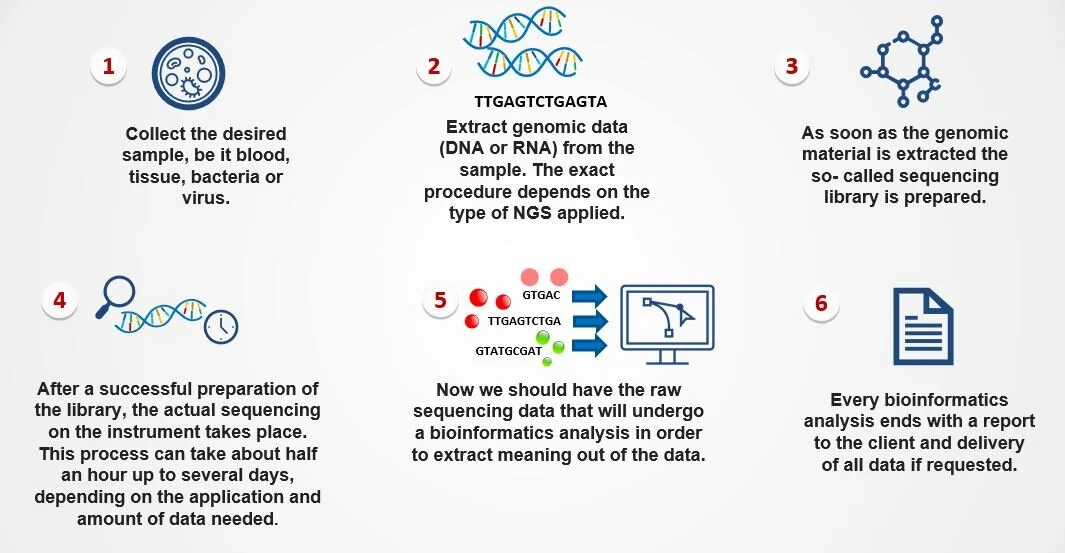
The principle steps when performing an analysis based on NGS are comparable for most cases:
Basic steps in NGS
2) What progress have you seen in bioinformatics in recent years?
A recent development is the aggregation of huge amounts of genomics data in private as well as in public data repositories like the European Nucleotide Archive and others. This development allows new types of comparative analyses, like studying the variation in the human genome and linking it to diseases. Without efficient and proper (bioinformatics) solutions a lot of the data will just be deposited without much use.
Michael Schmid, Founder of Genexa (www.genexa.ch)
Genexa has developed the nucleotID solution, which can be used to develop diagnostic tests
This wide availability of genomics data leads to the development of our newest product nucleotID which also is a result of several recent client projects in the field of pathogen diagnostics. We have been using this solution to help companies develop diagnostic tests.
A crucial first step for the development of a diagnostic molecular test is the identification of a suitable genomic target.
Our solution uses large amounts of public and proprietary genome information for the detection of ideal genomic markers and presents the results in an interactive graphical user interface for visual exploration – without the need for an in-house bioinformatician. To our knowledge we are the first start-up service provider for the discovery of genomic pathogen markers working in a fully automated fashion and making use of complete genome information.
How bioinformatics contributes to developing diagnostic tests
3) What are the main challenges you have faced in developing your bioinformatics solution and how have you overcome them?
First of all, nucleotID (Genexa’s bioinformatics solution) always has to deliver high quality results. Our customers want to be able to rely on our platform so that they can concentrate fully on the development of a new diagnostic test in the laboratory. So quality is key and most important.
In the context of nucleotID, high quality means that a reported genomic marker must meet customer defined requirements, but at the same time the workflow must not overlook potentially useful genomic markers. This can be ensured by rigorous testing after every change or extension. We also incorporate any client feedback if we see room for improvement. Before we deliver results, we will always perform a manual check as a last safeguard step to ensure high quality.
The second challenge was to fully automate the entire analysis process. Since we collect publicly available data from several databases, a lot of work was invested in the automated collection of data from remote locations in an efficient way. Once the collection step is complete, the public data can be combined with proprietary data from the client and the actual analysis can begin.
This will also be fully automated up to the generation of the final report. As you can imagine, when retrieving data from non-internal sources, one has to be prepared to deal with unexpected properties in the data.
Third, the results of nucleotID are quite complex and not easy to visualize in static reports and images. For this reason, we have developed a dedicated viewer that can be installed on the customer’s infrastructure and allows the visual inspection of the results – in addition to our text reports generated by nucleotID. This way the client, usually a biologist, can “play” with the results and understand them in more detail and intuitively.
4) What are some future trends you foresee in the next 5 to 10 years for NGS and bioinformatics, especially in terms of improving human health?
Clinical genetics is a field where we can expect massive benefits from implementation of current and new NGS technologies in the coming years. It involves the study and treatment of heritable disorders and disease predispositions.
Currently, applied screenings in this field often only examine a few well known candidate genes within the thousands of genes of the human genome. A large scale screening often is still too costly or too slow in many cases. This can prevent a quick and ideal treatment of the patients. The broader application of NGS in general and new NGS developments in particular will most likely change this and have a massive impact on clinical genetics and thus on the health of patients.
Bioinformatics not only has to enable the new applications that are emerging in clinical genetics but also has to be integrated into the daily workflows to allow the swift and easy analysis of disorders caused by genetic factors. So bioinformatics is an important factor for patient health, currently, but even more in the future.